Demography and genomics of outbreaking populations
Demography and Genomics of outbreaking populations
Demographic variation has major implications in spatial and temporal genetic diversity and structure, and consequently population genetic inferences, which are magnified in outbreaking systems. Thus, current models that aim to understand genetic dynamics of outbreaking populations through time and space risk making incorrect conclusions regarding landscape connectivity, dispersal capacity, and adaptation. The goal of this research theme is to challenge existing conceptual underpinnings of genetic variation, characterize how demographic context affects neutral and adaptive population genetic patterns, and to develop new approaches to address ecological, conservation, and management initiatives considering informative genetic and genomic perspectives. This work primarily focuses on outbreaking forest insect pests such as the eastern spruce budworm. However, long-lived species (e.g., turtles) also present complex demographic contexts that confound traditional approaches to population genetic inference.
Eastern spruce budworm
The eastern spruce budworm (SBW; Choristoneura fumiferana) is a lepidopteran forest pest that defoliates huge areas of spruce and fir forests during its periodic outbreaks, with major socio-economic implications. SBW outbreak dynamics are shaped by the complex interactions between climate, forest structure, competition, parasitism and predation, and dispersal. In pursuit of our research goals, we have developed robust sample and genomic libraries spanning more than a decade and covering the majority of the SBW distribution across North America, during different phases of an outbreak cycle. This project has several key themes that we are exploring to better understand the interplay between outbreak dynamics and genetic inferences:
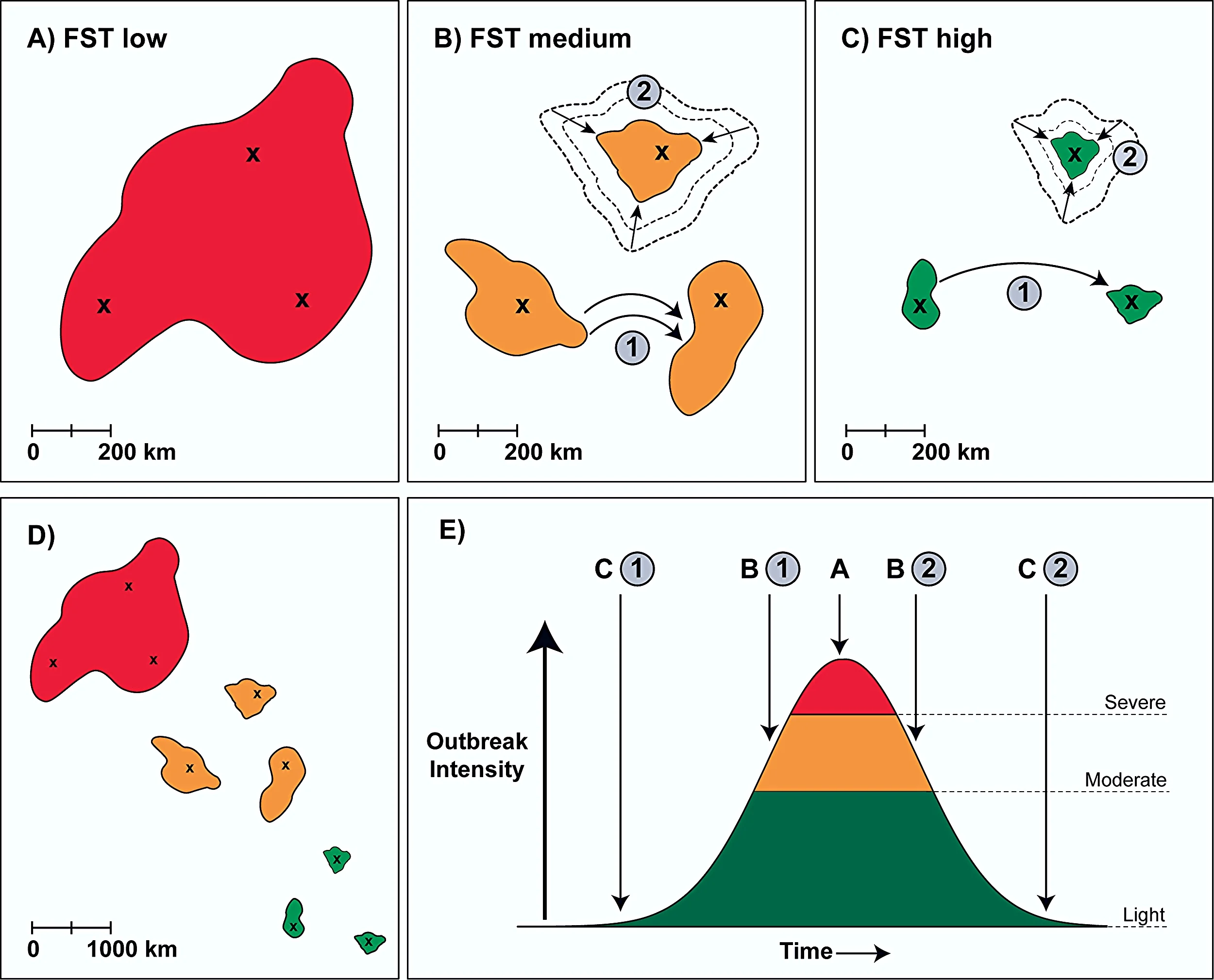
1. Despite the important role of dispersal to the spatial dynamics of SBW outbreaks, little is known about how it varies with spatial context and over the course of an outbreak, and how it affects spatial synchrony in outbreak dynamics. This research applies tools and methods from spatial population genetics to characterize genetic connectivity among outbreak patches in the current outbreak in eastern north America.
2. Using genetic information to infer connectivity and how it varies within and among years, we will infer patterns of gene flow and dispersal across the landscape, and the influence of intervening land-cover (isolation by resistance) and local environmental conditions (isolation by environment). Concurrently, we are developing dispersal models using predicted phenology (BioSIM) and demographic data collected from pheromone traps (adult males).
3. Spatial and temporal demographic variation in non-outbreaking populations has been the origin of many population genetic concepts, which have been the standard for developing metrics and analyses for practical applications such as conservation and management. Our research is challenging the understanding of how standard protocols for analysing population genetic data can lead to incorrect conclusions when investigating the rapidly changing, extreme demography of outbreaking dynamics.
4. We are developing efficient and robust forward in time, individual based simulation models to evaluate a range of demographic scenarios to better predict and manage outbreaking populations. Using our extensive real-world SBW data, we can test the accuracy of these simulations, with the goal of developing a tool that can be adapted to other outbreaking species and scenarios.
Together, this work will increase our knowledge of how SBW demographic dynamics vary in different landscape contexts. The results can be used to inform management initiatives and improve simulation models that predict insect population dynamics and forecast future outbreak risk. This work will also address the important question of the potential efficacy of currently proposed reactive and proactive (e.g. early intervention) management strategies.
Selected Publications
Fontenelle, J. P., Larroque, J., Legault, S., Wittische, J., Underwood, J. A. R., & James, P. M. A. 2024. Multiyear Genotype Characterization of Eastern Spruce Budworm Outbreaking Populations from Quebec and Adjacent Regions. Ecology , 105(12)
Larroque J, Johns R, Canape J, Morin B, James PMA. 2020. Spatial genetic structure at the leading edge of a spruce budworm outbreak: the role of dispersal in outbreak spread. Forest Ecology and Management 461, 117965
Larroque J, Legault S, Johns R, Lumley L, Cusson M, Renaut S, James PMA. 2019. Temporal variation in spatial genetic structure during population outbreaks: distinguishing among different potential drivers of spatial synchrony. Evolutionary Applications 12(10): 1931-1945
James PMA, Cooke B, Brunet B, Lumley L, Sperling FAH, Fortin M-J, Quinn V, Sturtevant BR. 2015. Life-stage differences in spatial genetic structure in an irruptive forest insect: Implications for dispersal and spatial synchrony. Molecular Ecology. 24(2): 296-309.

SBW larva hanging by a thread. Photo: Olivier Pontbriand-Paré